-
LungMAP3 Team
-
Cincinnati Children's HospitalView full details: Cincinnati Children's Hospital
Principal Investigators



Yan Xu, PhD, Principal Investigator Kathryn Brokamp- Wikenheiser, MD, PhD,co-Investigator 
Jeffrey Whitsett, MD, co- Investigator Research Description
A mechanistic 3D AI-powered map of genetic chILD
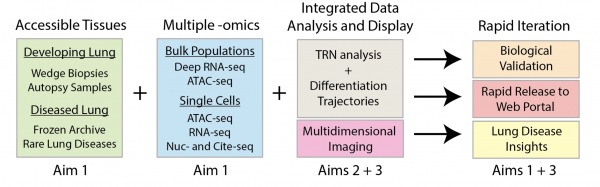
Leveraging the technologies and infrastructure in our previous LungMAP phases, we will construct a multi-scale map of developing acinar and alveolar structures during normal childhood and in the setting of childhood interstitial lung disease (chILD) characterized by aberrant lung morphogenesis. Linked to gene variants, chILD provides a unique opportunity to understand key genetic pathways and cellular interactions of human acinar and alveolar development, in turn informing injury-repair associated with lung diseases throughout the lifespan. ChILD has been linked to mutations in FOXF1, TBX4, NKX2-1, and surfactant genes and thus provides a unique opportunity to harness genetics to understand human lung acinar development. Unlike typical molecular phenotyping efforts that grapple with causal versus associative changes, our map of chILD will leverage our unique collection of samples with genetic alterations to construct mechanistic gene regulatory networks (GRNs) based on experimental and computational, including artificial intelligence (AI), evidence. Deciphering these GRNs will have impact beyond chILD by addressing the unmet medical need of interpreting genetic variants and identifying GRN members critical to acinar formation and maintenance in BPD, COPD, IPF, etc. Our focus on the acinus as the basic unit of the lung parenchyma, inclusive of respiratory bronchioles, alveolar ducts, alveolar sacs, and alveoli, tackles the proximal-distal heterogeneity in lung biology and diseases. Working in a collaborative LungMAP3 consortium, including other Research Centers, the Data Coordinating Center (DCC), the Human Tissue Core (HTC), our LungMAP3 project will use single-cell and spatial multiomics, high-resolution confocal and electron microscopy, patient-derived iPSCs, and bioinformatics including AI tools to define, model, and predict normal and diseased lung development. The resulting mechanistic insights and enabling technologies will be disseminated to the entire lung community via disease atlases and web portals, as we have done during LungMAP1 and 2, to accelerate lung biology discoveries and disease therapies. Our pipeline of defining (Aim 1), modeling (Aim 2), and predicting (Aim 3) diseases will illuminate acinar biology and establish enabling technology to benefit the whole lung community.
Cincinnati Children's Hospital Medical Center3333 Burnet Avenue, Cincinnati, OH, USAMLC 5041Cincinnati , OH 45229 -
University of PennsylvaniaView full details: University of Pennsylvania
Principal Investigators

Research Description
One of the major goals of LungMAP3 is to extend the growing knowledge of cellular heterogeneity within the human lung towards defining the spatial, molecular, and cellular changes that occur in human respiratory diseases. The ability to interrogate both rare and common lung disease will allow for the rapid advances of the molecular pathogenesis of these diseases and identify novel therapeutic targets. Findings from such studies will need further validation using new models of human lung cell biology including organoids and ex vivo explants. This will entail the acquisition of human adult lung samples, both normal and diseased, extending beyond what has already occurred as part of the LungMAP Consortium. The University of Pennsylvania LungMAP Center will focus on three emerging concepts and challenges: 1) accurately and precisely phenotype adult human lung diseases at the spatial and single cell level with a focus on COPD, ILDs and more rare diseases including alpha-1 anti-trypsin deficiency (A1AT), 2) identify molecular defects in progenitor cell niches in human chronic lung diseases (CLDs) through dynamic integration of single cell analytics with spatial resolution technologies to elucidate disease progression signatures, and 3) develop and implement new model assay systems to mechanistically define molecular and cellular defects during the progression of CLDs. This will require the use of advanced transcriptomic, epigenomic, and bioinformatic approaches to phenotyping cell-cell communication and cell-niche interactions. The Penn LungMAP Research Center has an existing pipeline to acquire normal and diseased lung tissue, and we have developed extensive informatic software to interrogate changes in cell fates and states in disease from deep spatial and single cell analysis. The Penn LungMAP Research Center has been extremely productive in LungMAP Phase 2 and has defined new cell lineages such as RASCs in the human lung, defined novel aspects of emphysematous disease pathology including COPD, and begun to define rare CLDs such as A1AT at a single cell level. The overarching strategy of the Penn LungMAP 3 Research Center is to phenotype the cellular and molecular changes that occur in CLDs at the single cell level and identify the mechanisms that drive CLD progression using novel approaches to integrate progenitor cell niche regulation using carefully validated ex vivo model systems.
University of PennsylvaniaUniversity of Pennsylvania 3400 Civic Center BoulevardBuilding 421Philadelphia , PA 19104 -
University of California San DiegoView full details: University of California San Diego
Principal Investigator



Xin Sun, PhD, Principal Investigator Jamie Verheyden, PhD, co-Investigator Laura Crotty Alexander, MD, co-Investigator Reseach Description
The central goal of the UC San Diego LungMAP III project is to map across multiple chronic lung diseases for shared and distinct signatures using cutting-edge multi-modal approaches. We will leverage such disease diversity to reveal rich biology. Questions include: 1) how many different types of transitional cells and myofibroblasts are there and how they differ from disease to disease; 2) where are the signaling hubs that drive pathogenesis; 3) do rare cell types change or emerge; 4) can neuroimmune signals be used to define disease-specific cellular communities. We will test the central hypothesis that different lung diseases can be distinguished by a unique set of signatures, including cell type/cell state composition and their spatial organization, transcriptomic and epigenomic profiles, and signaling and extracellular matrix dynamics. In Phase III, we will continue our productivity as in Phase II in producing a large number of datasets for open access by the research community. We will also continue our effort in contributing to resources such as Lung CellCards. We will continue to bring cutting-edge technologies, some developed by our partner at UC San Diego Center for Epigenomics, to the lung community. We expect that the datasets that we will generate in phase III will seed many paradigm-shifting studies by the collective lung research community.
University of California San Diego9500 Gilman DrLa Jolla , CA 92093 -
University of California Los AngelesView full details: University of California Los Angeles
Principal Investigators



Brigitte Gomperts, MD, Principal Investigator, UCLA Darrel Kotton, MD, co- Investigator, Boston University Katherin Plath, PhD, co- Investigator, UCLA 


Geremy Clair, co- Investigator, PNNL Sam Weigt co-Investigator, UCLA Nicholas Jackson, Collaborator, UCLA Research Description
Interstitial lung diseases (ILD) are a large group of lung diseases that are characterized by chronic, progressive pulmonary fibrosis. We hypothesize that in all ILDs the progressive fibrosis is driven by ongoing injury/stress in the airway epithelial cells that set up crosstalk with their neighboring mesenchymal cells resulting in a chronic wound healing process that alters the matrix and changes the cellular niche. The goal of the proposed research is to address how interactions between the epithelial, mesenchymal and matrix compartments drive progression of fibrosis in all forms of ILD. We are proposing to study these interactions between the epithelial, mesenchymal and matrix compartments, which is now feasible with spatial transcriptomics, single cell secretion protein analysis, and small region proteomics. Most studies have looked at end stage disease and fibrotic lesions where most of the epithelial cells are lost and consequently the initiating and propagating factors are not well understood. We propose to study Interstitial Lung Abnormality (ILA) lesions, which represent very early ILD lesions, early stage ILD lesions when patients start to become symptomatic, and late stage ILD regions that still contain epithelial cells so we can study their interactions with mesenchymal cells and matrix. We have assembled a team with expertise in tissue biobanking and metadata collection (Weigt and Jackson), spatial transcriptomics and ILDs (Gomperts), novel bioinformatics pipelines and integration of genomics data (Plath), regional small-scale proteomics in ILDs (Clair), iPSC-derived AT2 disease modeling and ILDs (Kotton) and bioengineered models of ILD (Gomperts).
We will use our biobank of ILD and ILA samples and prospectively collect ILD and ILA patient samples to profile the airway epithelium and underlying fibroblasts from bronchioles to distal airspaces with spatial small region transcriptomics and single cell spatial genome-wide transcriptomics. We will also use the secretory-single cell profiling (Sec-seq) technology to capture single airway epithelial cells, from fresh ILA and ILD tissue and trap the secretions from each cell to identify secreted factors. Nanoscale small region proteomics will be used to identify proteins in the adjacent epithelial, mesenchymal and basement membrane matrix compartments from ILA and ILD samples. To validate these findings and identify mechanisms by which persistent airway injury induces progressive fibrosis we will use cell co-culture models of iPSC derived AT2 cells with specific mutations associated with familial IPF, in conjunction with primary healthy or IPF lung fibroblasts. These powerful reductionist models allow the study of specific epithelial-mesenchymal-matrix interactions that drive progressive fibrosis.
UCLA6230 Wilshire Blvd Ste 2000Los Angeles , CA 90048 -
Harvard UniversityView full details: Harvard University
Principal Investigators



Benjamin Medoff, MD, Principal Investigator, Massachusetts General Hospital Jacques Deguine, PhD, co- Investigator, The Broad Institute Jay Rajagopal, MD, co-Investigator, Harvard University Research Description
Mapping Airway Epithelial and Immune Cell Interactions in Lung Health and Disease
Our project within LungMAP3 focuses on a comprehensive analysis of cellular communication within the airways in health and in asthma and chronic obstructive pulmonary disease (COPD). These diseases, which together account for the highest global burden of disability and mortality from chronic respiratory diseases, are characterized by altered interactions between airway epithelial cells (AECs) and immune cells, leading to chronic inflammation and structural remodeling of the airways, which progressively disrupts normal lung function.
We hypothesize that unique AEC-immune circuits, involving both common and rare AEC subtypes, as well as specialized structures like hillock islands and inducible bronchus-associated lymphoid tissue (iBALT), are central to driving the chronic inflammation seen in asthma and COPD. These structures appear to foster resident memory T cells and B cells, perpetuating local immune responses that shape the clinical trajectory of each disease. Our overarching goal is to construct a highly detailed, spatially resolved atlas of these interactions, which could pinpoint precise therapeutic targets for modulating disease-specific pathways in asthma and COPD.
Using airway samples from research bronchoscopies, lung resections, and explant tissue, we will employ single-cell RNA sequencing and high-resolution spatial transcriptomics to define the spatial arrangements and interactions between immune-responsive AECs and immune cells in three patient cohorts: asthma, COPD, and healthy controls. Further, using a 3D organoid system, we will also functionally validate these interactions by disrupting specific signaling pathways with CRISPR modifications, thereby isolating disease-relevant phenotypes associated with AEC-immune cell communication in asthma and COPD.
To capture the adaptive immune response in each condition, we will analyze airway brushings for T and B cell receptor diversity using single-cell TCR and BCR sequencing. Spatial mapping of these receptors in resections and explant tissue will allow us to identify and localize disease-driving immune clones that expand within affected tissues. This approach is expected to reveal disease-relevant specificities of T and B cell populations in each disease, uncovering distinct or shared immune responses that could inform targeted therapeutic strategies.
Through this research, we aim to delineate the distinct and overlapping cellular communication networks that contribute to chronic inflammation in asthma and COPD, which could lead to new precision therapies. Our findings will support the creation of a detailed cell-specific atlas of the airway immune microenvironment in health and disease, making valuable contributions to LungMAP3.
Harvard University25 Shattuck StreetBoston , MA 02115Mass General Hospital55 Fruit Street,Boston , MA 02114Broad75 Ames StCambridge , MA 02142 -
University of Rochester: LungMAP Human Tissue CoreView full details: University of Rochester: LungMAP Human Tissue Core
Principal Investigators



Gloria Pryhuber, MD, Principal Investigator Ravi Misra, Ph.D, co-Investigator Thomas J. Mariani, Ph.D, co-Investigator 

Phillip J. Katzman, MD, co-Investigator Gail H. Deutsch, MD, co-Investigator Research Description
In collaboration with:
National Disease Research Interchange
International Institute for the Advancement of Medicine
The research team at the University of Rochester Medical Center has served as the Human Tissue Core (HTC) for LungMAP since the program’s inception in 2014. The BioRepository for INvestigation of Diseases of the Lung (BRINDL) was established to preserve samples collected from predominantly non-diseased pediatric donors in collaboration with organ procurement organizations to obtain material authorized for research when not suitable for transplant. In LungMAP II, we expanded the repository to include pediatric rare diseases including bronchopulmonary dysplasia, children’s interstitial lung disease (chILD), pulmonary hypoplasia and the more common disease, asthma. BRINDL now contains over 480 human, non-fetal, lung sets spanning mid-late gestation to adult age, as well as an additional 90+ with specified disease. Sample has been utilized by approximately 60 investigators and institutions across the world to complete cellular, molecular, and microscopic studies, contributing to over 50 publications. In LungMAP Phase III, the HTC will continue to collect tissue using the established workflow. Because of the concern and complexity in recovering high-quality lungs in a standardized manner and with as little ischemia-induced artifact as possible, and the need to meet all legal and ethical standards, we continue to choose to work primarily through elements of the United Network of Organ Sharing, the federally contracted Organ Procurement and Transplantation Network (OPTN), and two non-profit organizations to whom are referred non-transplantable donated organs suitable for research, The National Disease Research Interchange (NDRI) and the International Institute for the Advancement of Medicine (IIAM). We are also available to Pediatric Pulmonary Centers for referral of rare lung disease samples. In addition, we will assist LungMAP Phase III Research Centers in developing methods to collect and catalog tissue that fall into their disease domain expertise. Cases will include healthy and diseased donors from diverse ethnic and racial backgrounds. The HTC will assist RC teams in collecting detailed metadata for each sample and such information will be included in the BRINDL database. Biospecimens from all phases of LungMAP will be available to qualified LungMAP and non-LungMAP researchers (https://brindl.urmc.rochester.edu), and integrated into the LungMAP Data Coordinating Center’s portal system. Our URMC-HTC, in collaboration with Seattle Children’s Research Institute, will prepare the donated organs and tissues to meet the needs of the LUNGMAP Phase III. Finally, the BRINDL repository will be prepared for long-term access as the LungMAP program sunsets at the end of Phase III.
BRINDL can be accessed from here.
URMC
University of Rochester Medical Center
School of Medicine and Dentistry
601 Elmwood Ave, Box 850
Rochester , NY 14642
University of Rochester Medical CenterUniversity of Rochester 601 Elmwood AvenueRochester , NY 14642 -
Vanderbilt UniversityView full details: Vanderbilt University
Principal Investigators


Jonathan A. Kropski, MD, Principal Investigator Jennifer Sucre, MD, Principal Investigator, Vanderbilt Nicholas E. Banovich,PhD, Principal Investigator, The Translational Genomics Research Institute Research Description
Vanderbilt University Medical Center/Translational Genomics Research Institute LungMAP3.0 Research Center
Single-cell genomic studies have transformed our understanding of the cellular organization and plasticity of the human lung, providing substantial insights into molecular pathways active in end-stage lung diseases including pulmonary fibrosis (PF), bronchopulmonary dysplasia (BPD) and others. A common theme emerging across these studies is that there is a profound loss of highly specialized cells and cellular phenotypes in advanced lung diseases. The critical signals responsible for tissue organization and cellular specialization during lung development occur in three-dimensional space, and spatial context is critical to understand the intercellular communication mechanisms, and relationships among different cell types and states that establish and maintain distal lung “niche specialization”. These observations lead us to hypothesize that perturbations of cell-cell signaling in the alveolar niche lead to loss of cellular specialization, and disease-specific programs of progressive pathologic remodeling in PF, BPD and other chronic lung diseases. The goal of this LungMAP3.0 Research Center is to identify key molecular targets at the early stage of disease most amenable to stabilization or reversal. Our specific aims are to: 1) Develop a spatially-resolved molecular atlas of chronic parenchymal lung diseases, 2) Define mechanisms of niche dysregulation that result in the progressive histopathologic patterns of chronic lung diseases across the lifespan and 3) Investigate the transcriptional regulatory mechanisms through which niche-perturbations promote and prevent disease progression. We have assembled a team of investigators with a strong history of productive collaboration who offer expertise in chronic lung diseases that manifest across the lifetime and share a deep commitment to rapid and transparent data sharing in support of the lung biology community. We are excited to partner with other LungMAP Research Centers, the Data Coordinating Center and Human Tissue Core as we leverage state-of-the-art single-cell and spatial transcriptomic approaches that enable interrogation of archival samples from highly unique cohorts of early-stage lung disease as we investigate the molecular origins of alveolar niche dysregulation in IPF, other forms of interstitial lung disease, and BPD.
Vanderbilt University2301 Vanderbilt PlaceNashville , TN 37235TGen445 N. Fifth StreetPhoenix , AZ 85004 -
Data Coordinating CenterView full details: Data Coordinating Center
Principal Investigators

Benedict Paten, PhD, co-Investigator, University of California Santa Cruz
Research Description
The LungMAP 3 Data Coordination Center (DCC) serves as a nexus of LungMAP collective knowledge and activities. The DCC is responsible for data collation, re-analysis, and integration; secondary annotation tracking; developing tools to facilitate collection, sharing and data dissemination; operating a web resource for data, expertise, and collaboration; and coordinating activities across the Research Centers (RCs) and Human Tissue Core. The DCC also facilitate literacy for investigator use of developed tools and best practices for analysis, data provenance and metadata annotation, and engage the larger research community. To host the DCC, we have assembled a multidisciplinary team with data network leadership, along with leaders in single-cell genomics, image analysis, functional inference, and data re-utilization. The DCC leverages unique expertise at CCHMC, UCSC, and the Broad Institute to interoperate pulmonary-oriented single-cell and high-resolution imaging data with other atlas programs. We also include world-renowned pulmonary researchers into our leadership team to ensure the data and knowledge we provide to the research community has the greatest scientific impact. Collectively, we propose to accelerate the LungMAP scientific agenda by coordinating efforts across funded Centers, the NIH, and the pulmonary research community; cross-validate, annotate, deposit and link Consortium datasets and metadata that encompass advanced AI enabled tools, molecular -omics, spatial transcriptomics, imaging, and associated structural models; and enable sharing of data, results, and models within LungMAP and the research community. The lung disease datasets and results derived from the RCs are expected to yield significant new insights into intra-donor variation and disease pathogenesis. To ensure the underlying data produced by the RCs is findable, accessible, interoperable and re-usable (FAIR), the DCC will work closely with the RCs to establish and share best practices, coordinate metadata annotation, ensure studies are sufficiently powered, assist with the deposition of harmonized data of high integrity to secure repositories, and provide data access and standardized analysis workflows. Through the continued development of structured ontologies and metadata frameworks, RC-derived datasets will be annotated and harmonized using emerging best practices. The DCC will support the ingestion and validation of data and analysis from new technologies as they emerge. We will support the generation of centralized, cloud-enabled data processing workflows that are compatible with external initiatives such as HubMAP, BRAIN, and the HCA. Providing these functions in a web-enabled LungMAP Commons will promote interaction across many stakeholders.
Cincinnati Children's Hospital Medical Center3333 Burnet Avenue, Cincinnati, OH, USAMLC 5041Cincinnati , OH 45229University of California Santa Cruz1156 High StreetSanta Cruz , CA 95064Broad Institute105 BroadwayCambridge , MA 02142
-
-
LungMAP 2 Team
-
Cincinnati Children's Hospital Medical CenterView full details: Cincinnati Children's Hospital Medical Center
Principal Investigators

Jeffrey A. Whitsett, M.D. (Contact PI)
Cincinnati Children's Hospital Medical Center
Yan Xu, Ph.D. (Co-PI)
Cincinnati Children's Hospital Medical CenterResearch Description
Building a Multidimensional Map of Developing Human Lung – Jeffrey A. Whitsett, M.D. PI
Our LungMAP2 project seeks to work in a collaborative consortium to develop operational pipelines to study human lung development from the saccular, alveolar stages of morphogenesis at single cell level of resolution. The project will integrate single cell transcriptomic, chromatin accessibility, 2D and 3D high-resolution confocal microscopy, immunofluorescence, multiprobe RNA in situ hybridization studies to create a detailed molecular map of the cells, cell-cell and cell scaffold interactions in their precise anatomic positions in the developing human lung. Our LungMAP2 Center will work in close concert with the Data Coordinating Center (DCC), other LungMAP2 Centers and the Human Tissue Procurement Core (HTPC) to obtain, annotate, preserve and store human tissues using optimal conditions for imaging and omics analyses. Our LungMAP2 project will produce a detailed structural expression atlas of multiple regions, niches in the human lung. We will develop and validate new technologies for tissue clearing, 3D reconstructions of high-resolution images, multi-probe RNA and IF analysis. We will develop new tissue digestion single cell and single nuclear RNA analyses for precise regional niches of the normal developing human lung, tissue from and in infants with rare congenital lung disorders. We will work in a Consortium to develop useful bioinformatic programs, modeling cell-cell, cell matrix interactions to define the signaling and transcriptional programs building and maintaining organ structure and function. Working with the DCC and other LungMAP2 Centers, our bioinformatics pipelines will enable data visualization, interrogation in user friendly web portals for sharing with members of the LungMAP2 Consortium, the DCC and for the research community in general.
 CCHMCCincinnati Children's Hospital Medical CenterDivision of Pulmonary Biology3333 Burnet AveCincinnati , OH 45229-3039
CCHMCCincinnati Children's Hospital Medical CenterDivision of Pulmonary Biology3333 Burnet AveCincinnati , OH 45229-3039 -
University of RochesterView full details: University of Rochester
Principal Investigators

Principal Investigator: Gloria S. Pryhuber, M.D.
Co-Investigators: In collaboration with: Tom Mariani, Ph.D. National Disease Research Interchange Ravi Misra, Ph.D. International Institute for the Advancement of Medicine Philip Katzman, M.D Gail Deutsch, M.D. (U of Washington, Seattle) Research Description
Biorepository for Investigation of Diseases of the Lung (BRINDL) - Phase II
Lung disease originating in the period between the late canalicular stage to mature alveolarization and peak lung function, between limits of viability and early adulthood, remains a major cause of morbidity and mortality. Research to discover novel treatments for these disorders is limited by the rarity of access to normal and diseased human lung across this age span. Having, as the original Human Tissue Core (HTC) for the Molecular Atlas of Lung Development Program (LUNGMAP), created the BioRepository for INvestigation of Diseases of the Lung (BRINDL), the University of Rochester Medical Center (URMC) submits this application in response to RFA-HL-19-021 to continue to serve as the HTC for LungMAP Phase 2 (LUNGMAP II). BRINDL now contains over 300 human full lung sets spanning mid-late gestation to adult age, as well as an additional 70+ with specified disease. It will continue to be URMC-HTC responsibility to sufficiently identify and manage tissue sources to meet the overall Program goal to build an open-access, comprehensive, molecular atlas of the late- stage developing and maturing lung. As the HTC, URMC will collect, process, deposit, and distribute to the four LUNGMAP Research Centers (RCs), and ultimately the greater research community, human lung samples representing late gestation through early adulthood in forms that will support temporal and spatial characterization of molecular profiles of functionally or anatomically defined cell types in the developing lung. In LungMAP II, the URMC-HTC supports the expansion of LUNGMAP to 25 years of age and to include neonatal and pediatric rare lung disease as well as normal lung development. Because of the concern and complexity in recovering high-quality pediatric lungs in a standardized manner and with as little ischemia-induced artifact as possible, and the need to meet all legal and ethical standards, we continue to choose to work primarily through elements of the United Network of Organ Sharing, the federally contracted Organ Procurement and Transplantation Network (OPTN), and two non-profit organizations to whom are referred non-transplantable donated organs suitable for research, The National Disease Research Interchange (NDRI) and the International Institute for the Advancement of Medicine (IIAM). NDRI and IIAM work well with the OPTN and with families who approach them directly during a pregnancy or illness expected to be lethal. In addition, we have contacts with Pediatric Pulmonary Centers around the country to identify potential sources for rare lung disease samples. Our goal is to provide an additional 250 lung set donations, or at minimum biopsies, for inclusion in the LUNGMAP BRINDL biorepository. Our URMC-HTC, in collaboration with Seattle Children’s Research Institute, will prepare the donated organs and tissues to meet the needs of the LUNGMAP Phase II RCs.
BRINDL can be accessed from here.
URMCUniversity of Rochester Medical CenterSchool of Medicine and Dentistry601 Elmwood Ave, Box 850Rochester , NY 14642 -
University of PennsylvaniaView full details: University of Pennsylvania
Principal Investigators

Edward E Morrisey, Ph.D. (PI)
University of PennsylvaniaResearch Description
Multi-Modal Characterization of Three Human Lung Niches at the Single Cell Level
The respiratory system is architecturally complex and comprised of many compartments or niches responsible for unique functions during respiration. While the human respiratory system exhibits a significant level of similarity with rodents such as mice, it contains unique compartments and structures that are poorly understood but likely to be important in understanding disease etiology and progression. As an example, the heterogeneity along the proximal-distal axis of the human airway is significantly different than in the mouse, which may underlie the lack of appropriate rodent models for many human lung diseases. This lack of understanding is similar for the human pulmonary vasculature, where few animal models of diseases such as pulmonary hypertension exist. A detailed analysis of these compartments and others in the developing human lung will result in the identification of new cell lineages and molecular signatures of individual cells across the proximal-distal axis of the airways and along the pulmonary vasculature. These data will need to be coupled with high resolution imaging techniques to build a cellular atlas of the developing human lung. One of the major goals of Phase 2 of the LUNGMAP Consortium, which was originally initiated in 2014, is to define the unique architectural, cell, and gene expression complexities of the developing human lung using sophisticated and emerging technologies including single cell analytics. Given the spatially specific architectural complexities of the human lung, we propose to focus on three important compartments or niches: 1) the proximal airways, 2) the distal airways and alveolus including the terminal and respiratory bronchioles (TBs and RBs), and 3) the pulmonary vasculature. We will utilize multi-modal genomic, epigenomic, and proteomic techniques to define the cellular and molecular heterogeneity in these three niches at the single cell level, and disseminate this information to allow investigators to extract cell-cell crosstalk that defines and maintains these three niches in the developing human lung. Our group has developed and applied novel genomic and imaging tools and designed interactive web applications to display and interrogate multi- dimensional data that allows for specific, interactive, and continuous ongoing analysis of the data generated in the LUNGMAP Consortium. Importantly, our group has demonstrated the ability to define cell-cell interactions within specific lung niches by integrating genomic data with high resolution imaging. The ultimate goals of our proposal are to 1) identify and map the cell lineages within three critical niches of the developing human respiratory system, 2) define their spatial organization in relation to each other, 3) provide novel datasets to allow researchers to identify the cell-cell interactions that are critical for their postnatal development, and 4) organize and display the data for broad access throughout the scientific community using multi-dimensional genomic and proteomic analysis tools.
UPennSmilow Center for Translational Research11-179, Bldg 4213400 Civic Center BlvdPhiladelphia , PA 19104 -
University of California, San DiagoView full details: University of California, San Diago
Principal Investigators

Principal Investigator: Xin Sun, Ph.D.
Research Description
Decoding the Cellular Niches Critical for Lung Maturation and Pathogenesis
The human lung is vast organ with exquisite regional differences in the form of cellular niches best defined at single-cell resolution. Emerging evidence led to growing appreciation that the collective roles of these fine specializations are fundamental for lung function. Guided by the scientific premise that these niches are often sites of pathogenesis, we propose to map these niches in normal and disease tissues and directly compare them at the single-cell resolution. This goal is directly responsive to the mission of LungMAP Phase 2 to “extend to more specific and rare cell types.” To achieve this goal, we have gathered an interdisciplinary team of established and rising, basic and clinical investigators with complementary strengths in developmental biology (Sun), cell biology and immunology (Prince, Sajti), technology development and informatics (Ren, Preissl, Wang and Xu). Together, we have a track record of studying the control of normal lung maturation, as well as mechanisms underlying a number of LungMap-highlighted diseases, including bronchopulmonary dysplasia, congenital diaphragmatic hernia, childhood interstitial lung disease, and pediatric pulmonary hypertension (PPH). We will use cutting-edge technologies such as single-nucleus ATACseq, single-cell and single-nucleus RNAseq, multiplex antibody and RNA in situ hybridization, and three-dimensional imaging. Using these, we will systematically define the exciting kaleidoscope of cell types in the human lung, with an emphasis on cellular niches that are central to pathogenesis.
UCSDUniversity of California, San DiagoCellular and Molecular MedicineEast 10879500 Gilman DrLa Jolla , CA 920092 -
Battelle Pacific Northwest National LaboratoryView full details: Battelle Pacific Northwest National Laboratory
Principal Investigators

Joshua N. Adkins, Ph.D. Contact PI
Pacific Northwest National Laboratory
James Paul Carson, Ph.D. mPI
University of Texas at Austin
Geremy Clair, Ph.D. mPI
Pacific Northwest National LaboratoryResearch Description
Research Center for Spatiotemporal Lung Imaging And Omics
Lung development is a complex process that is spatiotemporally coordinated by a myriad of molecular events occurring in the different microenvironment and cell types composing in the pulmonary tissue. In human this process is continued postnatally with the alveologenesis occurring during the first few months to year of life. Alveologenesis is a dynamic, coordinated process that requires the accurate spatial and temporal integration of signals to develop the intricate alveolar structure. While important progress has been made, significant knowledge gaps remain in our understanding of the molecular mechanisms driving postnatal development. For this reason, our NHLBI established Research Centers (RCs) to create a spatiotemporal molecular atlas of the developing lung (LUNGMAP) focused from birth up to early childhood (~8 years). In the first phase of LungMAP, our RC developed resources that enabled a much more detailed understanding of normal lung development up to early childhood. In LUNGMAP Phase 2, we will apply our successful approaches and our newest technologies to extend analysis of lung development into early adulthood as well as human disease; focusing on bronchopulmonary dysplasia (BPD), the most common morbidity of preterm infants which is characterized by delayed or deficient lung maturation. Within the lung, the relationship between space, anatomy, and function is fundamental. Therefore, our approach includes new unbiased 3D quantitative imaging approaches implemented with high spatial resolution, as well as cell-specific omics: proteomics, lipidomics, and metabolomics. The integration of these complementary data collection methods facilitates the establishment of a cell-specific spatial atlas with an incredible breadth of molecular profiles across the developing lung in normal and disease states.
Specifically, we will accomplish our goal of an integrated molecular atlas of lung development through the following aims: (1) Spatial imaging for a molecular atlas of the human lung in normal and diseased states, (2) Cell-specific omics for a molecular atlas of the human lung in normal and diseased states, and (3) Managing data to facilitate collaboration and data integration.
Overall, these aims will create unprecedented multi-scale browsable quantitative three-dimensional “Maps” of proteins, lipids, and metabolites across the developing lung, providing for many novel insights toward understanding both normal human lung biology and disease pathogenesis.
PNNLPacific Northwest National LaboratoryP.O. Box 999Richland , WA 99352UT AustinTexas Advanced Computing CenterAdvanced Computing Building (ACB)J.J. Pickle Research Campus, Building 20510100 Burnet Rd (R8700)Austin , TX 78758 -
Data Coordination CenterView full details: Data Coordination Center
Principal Investigators

Bruce J. Aronow, Ph.D. (Contact PI)
Cincinnati Children's Hospital Medical Center
Nathan Salomonis, Ph.D. (Co-PI)
Cincinnati Children's Hospital Medical Center
Timothy Tickle, Ph.D. (Co-I)
Broad Institute of MIT and HarvardBenedict Paten, Ph.D. (Co-I)
University of California Santa CruzResearch Description
The LungMAP 2 initiative will create detailed molecular maps of the neonatal, pediatric and early adult human lung to enable improved understanding of functionally and anatomically defined cell types. The Data Coordination Center (DCC) will serve as the nexus of LungMAP 2's collective knowledge and activities. The DCC is responsible for data collation, re-analysis, and integration; secondary annotation tracking; developing tools to facilitate collection, sharing and data dissemination; operating a web resource for data, expertise, and collaboration; and coordinating activities across the Research Centers (RCs) and Human Tissue Core. The DCC will also facilitate literacy for investigator use of developed tools and best practices for analysis, data provenance and metadata annotation, and engage the larger research community. To host the DCC, we have assembled a multidisciplinary team with data network leadership, along with leaders in single-cell genomics, image analysis, functional inference, and data re-utilization. The DCC leverages unique expertise at CCHMC, UCSC, and the Broad Institute to interoperate pulmonary-oriented single-cell and high-resolution imaging data with other atlas programs. We also include world-renowned pulmonary researchers into our leadership team to ensure the data and knowledge we provide to the research community has the greatest scientific impact. Collectively, we propose to accelerate the LungMAP scientific agenda by coordinating efforts across funded Centers, the NIH, and the pulmonary research community; cross-validate, annotate, deposit and link Consortium datasets and metadata that encompass molecular -omics, imaging, and associated structural models; and enable sharing of data, results, and models within LungMAP and the research community. The datasets and results derived from the RCs are expected to yield significant new insights into lung maturation, intra-donor variation and disease pathogenesis. To ensure the underlying data produced by the RCs is findable, accessible, interoperable and re-usable (FAIR), the DCC will work closely with the RCs to establish and share best practices, coordinate metadata annotation, ensure studies are sufficiently powered, assist with the deposition of harmonized data of high integrity to secure repositories, and provide data access and standardized analysis workflows. Through the continued development of structured ontologies and metadata frameworks, RC-derived datasets will be annotated and harmonized using emerging best practices. The DCC will support the ingestion and validation of data and analysis from new technologies as they emerge. We will support the generation of centralized, cloud-enabled data processing workflows that are compatible with external initiatives such as HubMAP, BRAIN, and the HCA. We expect that providing these functions in a web-enabled LungMAP Commons will promote interaction across many stakeholders. This will position the LungMAP DCC to become a hub for data sharing, data integration, collaboration and hypothesis generation for investigators studying lung development and disease.
DCC- CCHMCCincinnati Children's Hospital Medical Center3333 Burnet AveCincinnati , OH 45229-3039DCC- BroadThe Broad Institute of MIT and Harvard415 Main StCambridge , MA 02142DCC- UCSCBaskin School of Engineering1156 High StSanta Cruz , CA 95064
-
-
NHLBI
-
NHLBIView full details: NHLBI
Program Officer

Qing "Sara" Lin, Ph.D.
Role
NHLBI will have substantial programmatic involvement that is above and beyond the normal stewardship role in awards.
- NHLBI will attend and participate as a voting member in all meetings of the Steering Committee.
- NHLBI will serve as a liaison, helping to coordinate activities among and for the awardees, including acting as a liaison to the NIH, and as an information resource for the awardees about lung development research activities.
- NHLBI will review the scientific progress of the LungMAP consortium, access for compliance with operating policies developed by the Steering Committee, and may recommend to the NHLBI to withhold support, suspend, or phase out an award for lack of scientific progress or failure to adhere to policies established by the Steering Committee.
-
-
LungMAP 1 Team
-
Cincinnati Children's Hospital Medical CenterView full details: Cincinnati Children's Hospital Medical Center
Principal Investigators

Jeffrey A. Whitsett, M.D. (Contact PI)
Cincinnati Children's Hospital Medical CenterS. Stephen Potter, Ph.D.
Cincinnati Children's Hospital Medical CenterResearch Description
Our research plan focuses to the generation and analysis of detailed NexGen gene expression data for both human and mouse lung from the late canalicular-saccular through the alveolar periods of development. The lung is comprised of diverse parenchymal- and hematopoietic derived cells, whose abundance, differentiation, and functions vary developmentally, regionally, and among species. Temporal and spatial interactions among diverse cell types orchestrate formation and function of the lung. While many of the genes and networks regulating lung formation are shared among vertebrate species, the physiology, structure and regional specific cell types, and gene expression patterns vary between murine and human lung. To effectively translate data from mouse models to humans, it is critical to understand their molecular similarities and differences at the cellular level. We will generate a detailed cell specific RNA database for human and mouse lung parenchyma. Single cell RNA-seq will be interpreted with data derived from laser capture microdissection (LCM), FACS, RNA analyses, and molecular imaging to create an expression map of human and mouse lung. Computational and "systems biology" approaches will integrate these data to create readily accessible databases that will synergize research related to pulmonary development, disease, and function. This application will focus to the analysis of cells from normal lung parenchyma (e.g., conducing airway and alveolar epithelial cells; vascular endothelial cells, including venous, arterial, lymphatic, and capillary cells; stromal and adventitial cells, including fibroblasts, pericytes, myofibroblasts, and cartilage; and neuroepithelial and neuronal cells).
Aim 1: Cell Specific NexGen RNA Analyses to Create an "Atlas" of Gene Expression In the Murine Lung. Single cell isolation, FACS purification, and LCM of lung parenchymal cells, and RNA-Seq will create a temporal-spatial map of gene expression in the mouse from E15.5 to postnatal day 28.
Aim 2: Cell Specific NexGen RNA Analysis to Create an Atlas of Gene Expression In Human Fetal Lung. Single cell isolation, LCM, and FACS analysis followed by RNA-Seq will create a map of the temporal-spatial patterns of the gene expression during human lung formation from 22 weeks of gestation through the postnatal-alveolar period.
Aim 3: Bioinformatics Analysis of NexGen RNAs to Define Gene Expression During Murine and Human Lung Formation. Bioinformatics and computational biology of RNA sequence data from the murine and human lung studies will be analyzed, curated using defined anatomic ontologies, and integrated to provide a sensitive and quantitative measure of gene expression patterns of the precise RNA splice forms, microRNAs and other noncoding RNAs expressed during lung formation.
CCHMCCincinnati Children's Medical CenterDivision of Pulmonary Biology3333 Burnet AveCincinnati , OH 45229-3039 -
Children's Hospital Los AngelesView full details: Children's Hospital Los Angeles
Principal Investigator
David Warburton, M.D.
Children's Hospital of Los AngelesResearch Description
The following Goals are designed to achieve the overarching Strategic Goal to construct a rich multiscale atlas of human alveolar development and to readily share the resulting materials with other research centers in the UO1 and with the broad research community.
Goal 1. Make a digital map of alveolar development over time in vivo and in tissue pieces using novel micro- CT (uCT), micro MRI (uMRI) and Phase contrast X-ray (PCX) technology validated in mice and then in human lung tissue.
Goal 2. Make a digital map of spatiotemporal gene expression during alveolar development over time using newly modified high throughput multiplex ISH with novel "slice and dice", Vibra-SSIM confocal technology, in mouse and human lung tissue.
Goal 3. Make a digital map of the fine structure of alveolar matrix during development in mouse and human tissue.
Goal 4. Develop image-processing technology to meld digital multiscale images of alveolar structure with cell autonomous gene expression with extracellular matrix protein configuration into a readily navigable, annotated novel data resource for the scientific community.
SOPs will be developed and process driven milestones will be set up to ensure deliverability and return on this significant scientific investment. Drs. Warburton, Shi and Driscoll are expert alveolar biologists while Drs. Fraser, Moats and Lansford are expert imaging scientists who are adept at recording, registering and assembling digital, multiscale combinatorial maps of organ development in Drosophila and Mouse and have invented many novel and highly innovative techniques to do so. We propose to translate our novel concepts on alveolar development derived in mice rapidly to studies in vivo in humans.
CHLAChildren's Hospital Los Angeles4650 Sunset BlvdLos Angeles , CA 90027 -
Pacific Northwest National Laboratory / University of Texas at AustinView full details: Pacific Northwest National Laboratory / University of Texas at Austin
Principal Investigators
Richard A. Corley, Ph.D.
Pacific Northwest National LaboratoryCharles K. Ansong, Ph.D. (Contact PI)
Pacific Northwest National LaboratoryJames Paul Carson, Ph.D.
University of Texas at AustinResearch Description
We have assembled a research team with the necessary experience to successfully establish a molecular atlas of the mouse and human lung based on spatial imaging and cell-specific -omic technologies. Within organs such as the lung, the relationship between space, anatomy, and function is fundamental. Therefore, our approach will include imaging techniques with high spatial resolution, as well as cell-specific -omics. The future correlation of these complementary data collection methods will facilitate the establishment of cell-specific spatial informatics across the developing lung. Specifically, we will accomplish our goal of an integrated molecular atlas of lung development through the following aims:
Aim 1: Spatial imaging for a molecular atlas of the developing mouse and human lung;
Aim 2: Cell type- specific omics for a molecular atlas of the developing mouse and human lung;
Aim 3: Manage data and metadata to facilitate collaboration and data integration. Overall, these aims will create the first spatial-temporal molecular atlas of the mammalian lung during alveologenesis, which in coordination with the other LungMAP centers will provide an unprecedented array of information about the healthy developing lungs in mouse and human. The novel imaging approaches and the suite of integrated pan-omics capabilities (i.e. proteomics, lipidomics, metabolomics and activity-based protein profiling) developed and available in a single laboratory at PNNL represents a unique strength of the Research Center.
PNNLPacific Northwest National LaboratoryP.O. Box 999Richland , WA 99352UT AustinTexas Advanced Computing CenterAdvanced Computing Building (ACB)J.J. Pickle Research Campus, Building 20510100 Burnet Rd (R8700)Austin , TX 78758 -
University of Alabama at Birmingham / Yale University / University of North Carolina, Chapel Hill/ University of California, San Diego / Carnegie Mellon UniversityView full details: University of Alabama at Birmingham / Yale University / University of North Carolina, Chapel Hill/ University of California, San Diego / Carnegie Mellon University
Principal Investigators

University of Alabama Birmingham
Yale School of Medicine
University of North Carolina Chapel Hill
Ziv Bar-JosephCarnegie Mellon University
Research Description: Alveolar DevMap
The overall objective of “Alveolar DevMAP” UAB-UNC-UCSD-Yale-CMU Research Center is to generate a compendium of the dynamic and regional changes in epigenetic marks, microRNA, mRNA and proteins that happen during alveolar septation, and use this compendium to generate a dynamic temporal regulatory model of normal alveolar septation
Aim 1: To identify changes in coding and non-coding RNAs during alveolar septation;
Aim 2: To determine changes in global DNA methylation during alveolar septation;
Aim 3: To identify the shifts in transcription factor and proteomic profile during alveolar septation;
Aim 4: To use, extend and validate our analytical tools to model dynamic signaling and regulatory networks activated in lung development that will be shared with other members of the consortium.
In all aims we will use samples acquired from laser capture microdissection (LCM) of developing alveoli or fluorescence activated cell sorting (FACS) of dispersed lung cells, collected at tight intervals to allow detailed analysis. Confirmation by quantitative immunohistochemistry and in-situ for transcripts will be included as well as some experimental validations. Samples from the Human Tissue Core (HTC) will be supplied Dr. Ambalavanan who will oversee LCM and FACS at UAB. Samples will be distributed to Dr. Kaminski (Yale) for miRNA and mRNA analysis, Dr. Hagood (UNC) for DNA methylation, and Dr. Mobley (UAB) for proteomics. Data integration and computational model development will be done by Dr. Bar-Joseph at CMU. The Alveolar DevMAP and computational models developed through these studies will identify regulatory "control points" in alveolar development. See entire DevMap Team below.
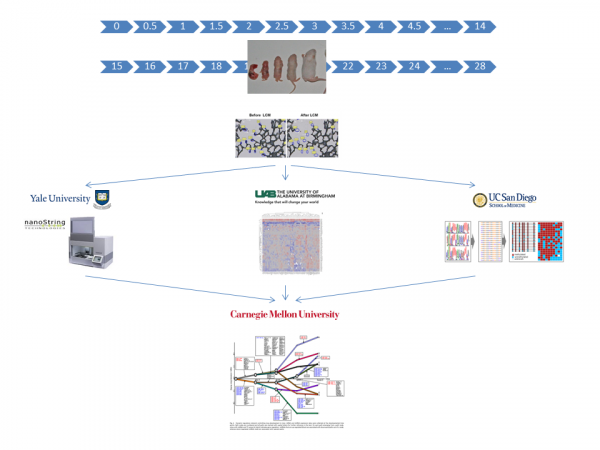 UABUniversity of Alabama at BirminghamDepartment of PediatricsLowder Building1600 7th Ave SouthBirmingham , AL 35233-1771YaleYale UniversityThe Anlyan Center300 Cedar St, STE S441DNew Haven , CT 06519UNCUniversity of North Carolina450 MacNider333 S Columbia St CB 7217Chapel Hill , NC 27599-7217Carnegie MellonCarnegie Mellon University5000 Forbes AvePittsburgh , PA 15213
UABUniversity of Alabama at BirminghamDepartment of PediatricsLowder Building1600 7th Ave SouthBirmingham , AL 35233-1771YaleYale UniversityThe Anlyan Center300 Cedar St, STE S441DNew Haven , CT 06519UNCUniversity of North Carolina450 MacNider333 S Columbia St CB 7217Chapel Hill , NC 27599-7217Carnegie MellonCarnegie Mellon University5000 Forbes AvePittsburgh , PA 15213 -
Human Tissue CoreView full details: Human Tissue Core
LungMAP Human Tissue Core: Biorepository for Investigation of Neonatal Diseases of Lung-Normal (BRINDL-NL)
University of Rochester Medical Center
Principal Investigator: Gloria S. Pryhuber, M.D.
Co-Investigators: In collaboration with:
Tom Mariani, Ph.D. Gail Deutsch, M.D. (U of Washington, Seattle)
Ravi Misra, Ph.D. Jeff Thomas (NDRI)
Jeanne Holden-Wiltse, M.S. Gina Dunne Smith (IIAM)
Philip Katzman, M.D.
Timothy Bushnell, Ph.D.
The Human Tissue Core (HTC) will procure, process, deposit and distribute normal late fetal, neonatal and early childhood human lung tissue and dissociated cells for LungMAP research working with United Network for Organ Sharing and their network partners.
LungMAP studies the biology of normal lung development, which makes tissue health a critical concern. We collect transplantation quality tissues through the International Institute for the Advancement of Medicine and the National Disease Research Interchange, organizations that link donors to the scientific community. A special effort, the Neonatal Organ Donation Program was established by IIAM to provide donations to LungMAP and other important research efforts. IIAM and NDRI receive referrals to families wishing to donate for research when transplantation is not an option.
At the HTC, donated lungs are:
- imaged by CT (computed tomography)
- reconstructed in 3D
- processed for histological analysis
- dissociated to provide a range of enriched cell populations including subsets of epithelial, endothelial, mesenchymal, lymphatic, immune and stem cells.
A working group of highly qualified Pathologists works with the HTC to:
- establish biorepository and assessment tools
- review specimens to assure quality and normality of the tissues chosen for LungMAP
We would like to thank
- The URMC Department of Pediatrics for support through the George Washington Goler Chair
- The URMC Department of Biostatistics and Computational Biology and The Center for Integrated Research Computing (CIRC) for computational support and development of inventory tracking systems using LabKey Software (https://labkey.com)
- NHLBI 1U01HL122700-01
Establishing a Neonatal and Pediatric Lung Donor Network
ELIGIBLE
•Live Born, either DBD or DCD (see next chart)•Ages: 20 weeks gestation to 10 years old•Known time of death and minimal warm ischemia time (WIT, preferred < 6, max 12 hrs)•Most commonly - lethal brain or cardiac disease, or for the older child, traumaEXCLUSIONS
•Unknown ischemic time or WIT > 12 hours•Primary lung injury (smoke inhalation, pneumonia, drowning)•Prolonged Ventilation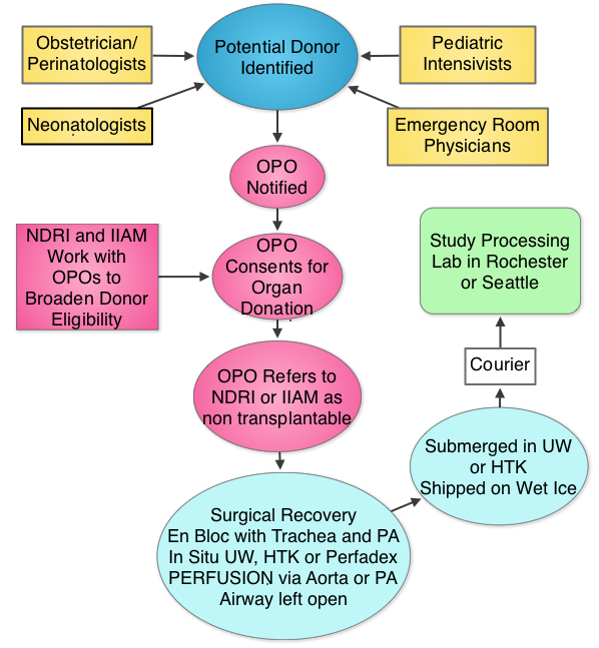 Human Tissue CoreUniversity of Rochester Medical CenterSchool of Medicine and Dentistry601 Elmwood Ave, Box 850Rochester , NY 14642
Human Tissue CoreUniversity of Rochester Medical CenterSchool of Medicine and Dentistry601 Elmwood Ave, Box 850Rochester , NY 14642 -
Data Coordinating CenterView full details: Data Coordinating Center
Principal Investigators
Scott M. Palmer, M.D. (Contact PI)
Duke UniversityRobert F. Clark, Ph.D.
RTI InternationalResearch Description
We have taken several innovative and cost-effective approaches to build the Data Coordinating Center (DCC) for the Molecular Atlas of Lung Development Program (LungMAP-DCC) that will greatly benefit the research community and promote a greater understanding of molecular lung development. We have created a multi-disciplinary team of investigators from Duke Clinical Research Institute, RTI International, and Cincinnati Children's Hospital Medical Center to lead and operationalize the LungMAP-DCC. We apply novel data management and bioinformatics approaches to create the Bioinformatics REsource ATlas for the Healthy lung (BREATH) database. The DCC performs data collection, integration, and analysis; develops and maintains the LungMAP database and website; and coordinates research activities of the Human Tissue Core (HTC) and the Research Centers (RCs). We complement these goals with the following Specific Aims that support the success of the consortium:
Aim 1: Administrative Coordinating Infrastructure. The DCC serves as the administrative infrastructure that facilitates collaboration among our multidisciplinary team, RC, and the HTC by:
- providing expertise in molecular lung development, clinical design, statistics, and bioinformatics; and
- the creation of consortium-wide priorities and policies, communication plan, and resource catalog; and
Aim 2: LungMAP Portal and BREATH (Bioinformatics REsource ATlas for the Healthy lung). The DCC is building the centralized data repository and public interface for LungMAP by creating and managing:
- BREATH;
- LungMAP Website;
- standard operating procedures for data management;
- existing lung development results;
- ontologies for lung development, structures, and cross species comparisons;
- experimental data from RCs and biologic sample data from the HTC; and
- novel tools to analyze the experimental data.
We leverage the complementary strengths of multi-disciplinary research teams with clinical pulmonary expertise, basic understanding of lung molecular development, successful leadership of multicenter research networks, and in-depth computer programing, database development, and bioinformatics skills that will create a highly functional, integrated, publically accessible platform that will facilitate analysis of data generated by the RCs and HTC to advance our understanding of molecular lung development.
DCC- DukeDuke University School of Medicine Office of the DeanDUMC 292740 Duke Medicine Circle124 Davison BuildingDurham , NC 27710DCC- RTIRTI3040 East Cornwallis RdP.O. Box 12194Research Triangle Park , NC 27709-2194
-
Consortium Centers





